Technical Summary eLife 2022
- E. E. Thomson, M. Harfouche, P. C Konda, C. Seitz, K. Kim, C. Cooke, S. Xu, R. Blazing, Y. Chen, W. S. Jacobs, S. Sharma, T. W. Dunn, J. Park, R. Horstmeyer* and E. A. Naumann*, “Gigapixel imaging with a novel multi-camera array microscope,” eLife 11, e74988 (2022) (*co-corresponding authors). Code, Data, Pre-Print, DOI, Project Page
Introduction:
Complex biological systems typically
include processes that span many levels of the organization across spatial
scales that vary by multiple orders of magnitude. Currently, no single imaging
system allows for unrestricted access to different scales simultaneously, due
to a lack of ability to jointly observe a very large field of view (the
observable area an imaging system can see) while maintaining a high resolution.
For example, many neuronal imaging methods typically require an animal to be
head-fixed or tethered, which restricts the natural behavioral repertoire and
inhibits spontaneous movement. Also, these methods can only follow one organism
at any given time, so it is hard to jointly observe the free movement and
interactions of many organisms at a high resolution over a large area.
Methodology:
To overcome these challenges, and extend observational capabilities to the gigapixel regime, we designed and constructed an inexpensive, scalable multi-camera array microscope (MCAM). This design is significantly cheaper than the other available large-area, high-resolution imaging systems as it leverages relatively inexpensive components. By adopting a parallelized approach to video acquisition, the MCAM removes a longstanding bottleneck of current single-camera microscope designs to provide researchers with greater flexibility, in particular for observing model organisms at high resolutions in an unconstrained manner.
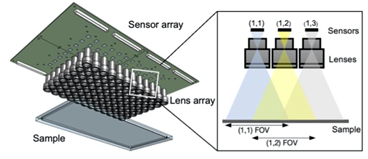
The setup in 3D
The frame rate of this first MCAM prototype was relatively slow (1 frame per second) and we have dramatically improved the frame rate since then. Please see our MCAM-54 and MCAM-48 websites for more information.
Results:
In this paper, we presented videos of multiple freely moving species, including Carpenter ants (Camponotus spp.), fruit flies (Drosophila melanogaster), nematodes (Caenorhabditis elegans), zebrafish (Danio rerio), and the amoeboid slime mold (Physarum polycephalum) over a 384 cm2 field of view. The MCAM allowed us to observe large-scale social behavioral patterns and morphological variations on spatial scales spanning multiple orders of magnitude.
Example video 2 – Carpenter ant behavior under multiple light/dark cycles
Additionally, the MCAM design permits stereoscopic tracking of the z-position of organisms by utilizing the overlapping field of view from adjacent cameras. This overlapping area guarantees that every point in the sample plane is observed by at least two cameras at the same time. This enables us to apply stereo depth measurement techniques by identifying positional disparities from matched characteristics and determine the depth of objects in the adjacent camera image pairs. To demonstrate the stereoscopic capability of MCAM, we traced the 3D path of zebrafish within the entire swimming arena.
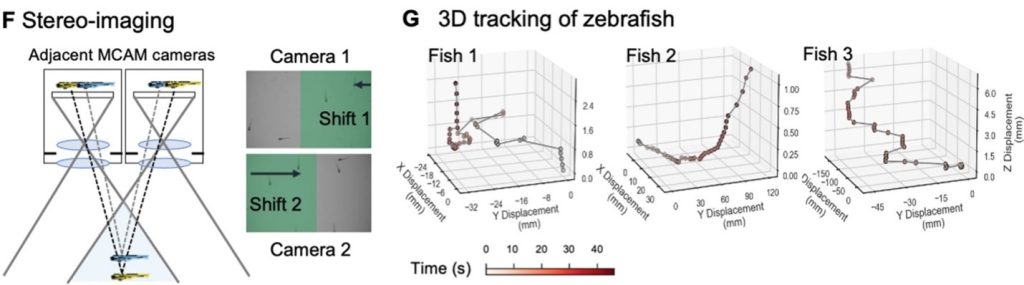
Conclusion:
Our new imaging technology can also be applied to a variety of other applications. Large-area, high-resolution imaging is important in industrial inspections of manufactured goods and materials, as well as in semiconductor metrology. For instance, such methods are required for defect detection on semiconductor wafer surfaces. It can also unlock new functionalities in imaging large pathology specimens, such as whole-mount brain slices. Biological researchers may also be able to utilize the MCAM platform to monitor bacterial colonies and cell cultures over macroscopically large areas.
