Gigapixel behavioral and neural activity imaging with MCAM
Affiliations:
- Duke School of Medicine Department of Neurobiology, Durham, NC 27710
- Biomedical Engineering, Duke University, Durham, NC 27710
- Ramona Optics Inc., Durham, NC 27701
Abstract
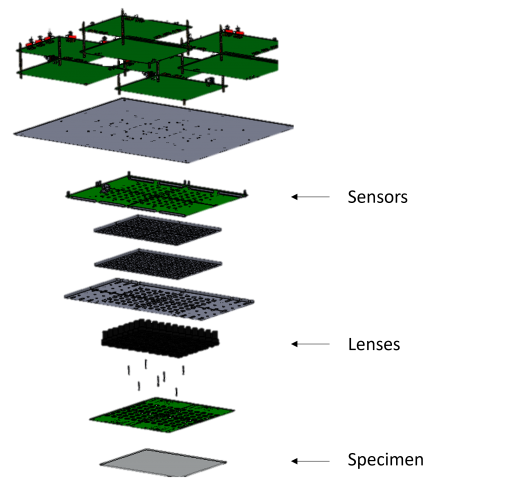
Overview of a Multi-Camera Array Microscope(MCAM)
Here we employ this system to observe behaviour and capture high resolution anatomical feature details for various freely moving model organisms over a large FOV. Thus, our system enables high-resolution recording from multiple spatial scales simultaneously which allows its usage in wide range of biological imaging applications, such as, studying cellular structures and capturing large-group behavioural dynamics .
We also demonstrate the ability to acquire dual channel fluorescence video of multiple freely moving zebra fish and record their neural activity using ratiometeric calcium imaging, which supplements the existing behavioural tracking analysis by providing more quantitative measurements for these moving specimen.
Example frame of freely swimming zebrafish larvae
Raw Video Snapshot
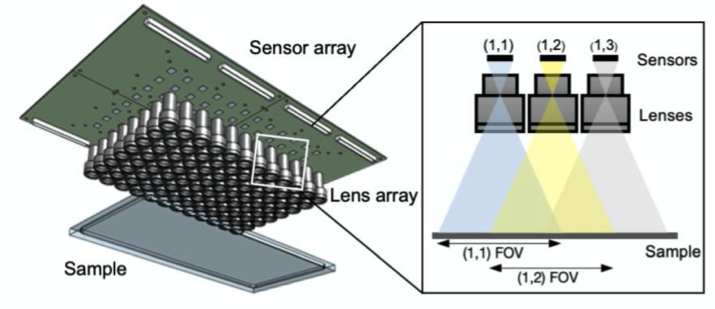
Gigapixel Imaging System: A.Four modular units in a 2×2 fashion, each consisting of 24 sensors, arranged together to demonstrate scalibility. B. Raw Video Acquisition
Design
Following are the design specifications of the acquisition system:
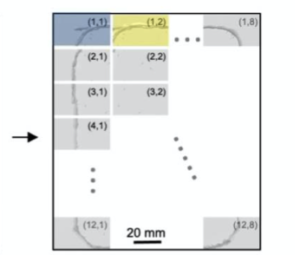
- The camera assembly consists of upto in 8 x 12 rectanglar array fashion
- FOV of imaging sample is 384 cm 2
- Full pitch optical resolution is 18 um
- Scalable
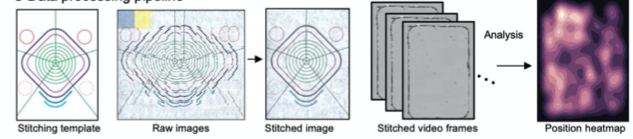
Generating image composite and performing analysis for MCAM
This is the heading
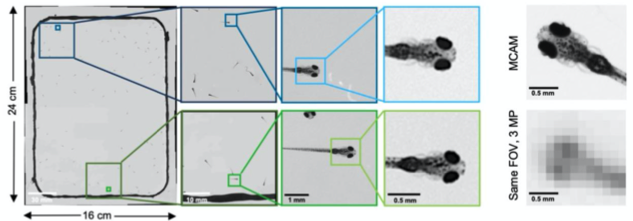
Observing Zebrafish behaviour at multiple spatial scales and comparing it with a single-lens system imaging same FOV(16x24 cm)
Methods
- Simultaneous Image acquisition from the camera array.
- Data acquisition post processing for efficient storage
- Standard image processing to correct for vignetting and brightness variations between the cameras.
- Digitally stitch acquired image into a single composite frame.
- Perform image analysis task such as organism trajectory assesment and multi-organism location measurement.
- Use machine learning and other computational algorithms to perform further analysis on the acquired dataset.
Results
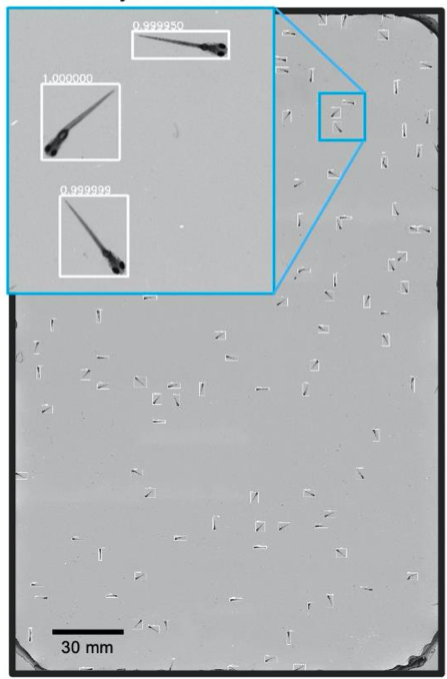
A. High-resolution, large-FOV imaging of large groups of larval zebrafish
- Recorded behaviour of ~100 wild-type 8 days post-fertilization zebrafish placed into a 22 x 11 3D printed arena.
- Resolved various morphological features of individual fish such as: pectoral fish position and distribution of melanophores.
- A CNN based object detector adopted to detect and crop individual organisms for further analysis
A. CNN Object Detection
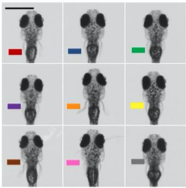
B. Zebrafish headshots
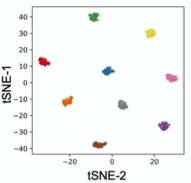
C. Zebrafish identification
D. Tail Tracking
E. Eye position tracking
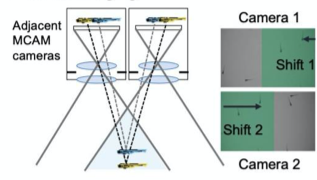
F. Stereoscopic Imaging and depth
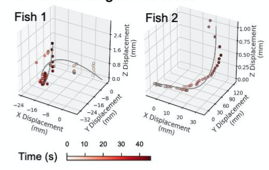
G. 3D. tracking of zebrafish, Z-displacement estimated by stereoscopic depth tracking
A) Nine Zebrafish headshots as input to the Siamese neural network for zebrafish identification. B) 2D Visualization demonstrating network capabilitiy to differentiate among the zebrafishes. D)Tail tracking visualization for three zebrafish E.) Histograms of Eye angles on 130 ZF across 20 frames F. Schematic of stereoscoic depth tracking.G.) 3D trajectory of 2 example larvae recorded gigapixel video of 130 WT zebrafish.
- Vast reduction in memory and dataset handling requirements was achieved by performing analysis only on the cropped images.
- Exploited high resolution imaging capability and differences in melanophore patterns to recognise individual zebrafish using morphological differences among the zebrafishes.
- Also identified behavourial details of each zebrafish by performing tail tracking and capturing eye position measurements.
B. Simultaneous behaviour and fluorescence imaging
- Integrated fluorescence imaging capabilities to MCAM unit by adding two sets of custom excitation sources for a 24 unit MCAM.
- Also mounted a bank of 24 emission filters via 3D printed filter array holder over each camera lens.
- The above addition enabled capturing epi-illuminated fluorescence images from each MCAM sensor.
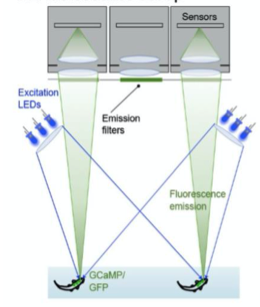
A. Fluoroscence Imaging setup
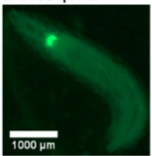
B. Drosophilla melanogaster example fluorescent MCAM image at high magnification
C. Calcium Imaging
D. Fluorescence Imaging
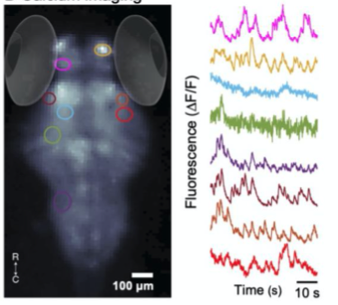
E. Neural activity monitoring during swimming for six example zebrafish captured in D.
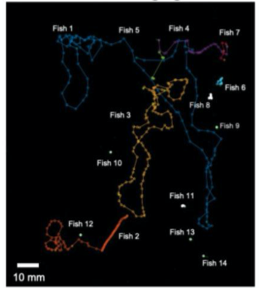
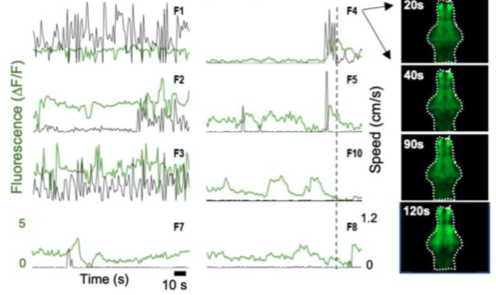
A. Fluorescence Imaging setup schematic, B. Drosophilla melanogaster example fluorescent MCAM image at high magnification C. Average fluorescent Imaging of agarose embedded zebrafish with, coloured sections represent ROI plotted on the right. Avg. ∆F/F traces demonstrate MCAM capabilities to resolve differences in neural activity, D. 14 Freely moving Zebrafish in a 24 camera MCAM system, tracked over a 120 s video, E. Fluorescent activities of six example zebrafish captured in D.
- Imaged freely moving drosophila larvae, epi-fluorescent setup enabled observation of proprioceptive neurons(responsible for movements in an individual) in the region of interests.
- Monitored neural activity by acquiring fluorescence images from zebrafish expressing GCaMP6( a genetically encoded fluorescent calcium indicator).
- Combined fluorescence and behavioral tracking, using a 24-camera MCAM, observed increase and decrease in neural activity marking onset and end of swimming bouts(seen in sporadically moving zebrafish).
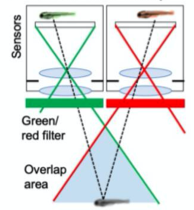
A. Dual Channel Setup
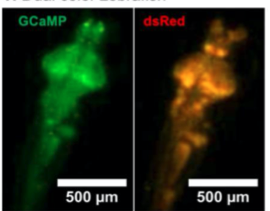
B. Dual Color Zebrafish
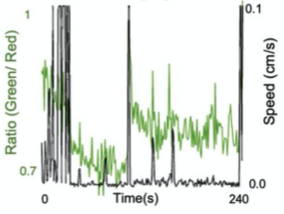
C. Ratiometric Imaging
A. Dual Channel Imaging configuration. Each area imaged by two MCAM sensors simultaneously, recording red and green fluorescence signals. B. Example dual-channel fluorescence image for a transgenic zebrafish expressing two different fluorescent signals indicators. C. Ratiometric analysis of neural activity for the sample example zebrafish. Movement is marked by increase in fluorescence signal.
- Installed alternating red and green fluorescent filters and utilized MCAM overlap between adjacent FOVs to perform ratiometric imaging.
- Captured dual channel fluorescence red signal doesn’t change and helps in compensating movement artifacts while recording GCaMP6 fluorescence
Example Fluorescence videos
Fluorescence activity of different Zebrafish at multiple spatial scales
